National College of Singapore physicists have demonstrated that a solitary guy-made molecular motor can exhibit a pressure related to by natural means transpiring types that electric power human muscle tissue. Their success are printed in Nanoscale.
Molecular motors are a class of devices with nanoscale proportions that are critical brokers of motion in dwelling organisms. They harness numerous electrical power sources within the physique to produce mechanical movement. A essential characteristic is the power created by a one motor in the course of its self-propelled movement. This force generation ability lets the molecular motor to deliver mechanical operate and is a measure of its electricity conversion performance, which influences its use in possible purposes.
The measurement of the force produced by normally transpiring molecular motors, which are usually produced of proteins, was realized two decades ago. On the other hand, similar measurements for artificial gentleman-built molecular motors produced of DNA (deoxyribonucleic acid) remain a problem. A study collaboration among the Molecular Motors Laboratory by Affiliate Professor Zhisong Wang and the Single-Molecule Biophysics Laboratory by Professor Jie Yan, the two from the Office of Physics, NUS has managed to detect the force generated by a relocating DNA molecular motor.
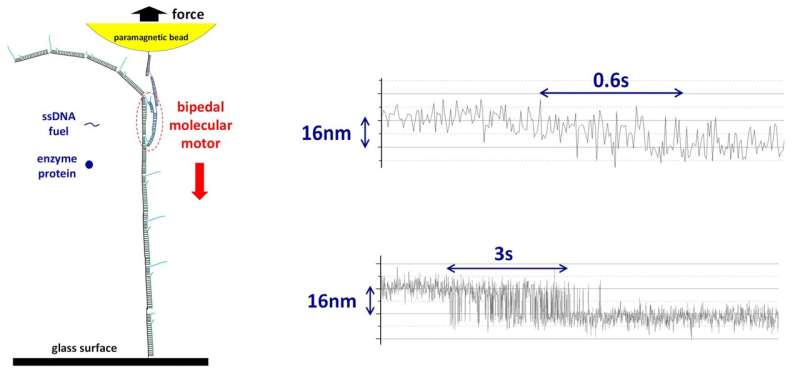
It is challenging to detect the forces created by a solitary molecular motor in movement for artificial motors because they are smaller and typically operate on smooth tracks (e.g., double-stranded DNA). Soft tracks are not fastened in position and are inclined to coil into a round condition. This affects the motion of the artificial motor. The analysis workforce overcame this problems by designing and executing in parallel one-molecule experiments that saved the tracks in position at the nanoscale stage though also concurrently detecting the miniscule power created by the going molecular motor. Using the magnetic tweezers procedure, they initial assembled an synthetic molecular motor and its track less than a paramagnetic bead (instrument for isolation of biomolecules). They then switched the paramagnetic bead to a drive detection manner (see Determine).
The investigate staff properly used their approach to an autonomous DNA molecular motor (previously developed by Prof. Wang’s lab). This bipedal molecular motor is ready to “stroll” consecutively on its own with a stride size of about 16 nm involving each and every phase, supplying a utmost force output of all over 2 to 3 pN. This measured force output is at a stage which is in close proximity to to in a natural way occurring molecular motors powering human muscle tissue, indicating a reasonably economical conversion of chemical vitality to mechanical motion.
Prof. Wang explained, “This review paves the way for the development of programs related with translational synthetic molecular motors which have to have the generation of forces. Examples incorporate molecular robots and biomimicking artificial muscle mass. Separately, the solitary-molecule process established in this function is relevant to force measurement of quite a few other artificial molecular motors with smooth tracks.”
Physicists create self-directed molecular motors that wander on tracks
Xinpeng Hu et al, Single-molecule mechanical study of an autonomous synthetic translational molecular motor past bridge-burning style and design, Nanoscale (2021). DOI: 10.1039/D1NR02296B
Citation:
Solitary-molecule experiments expose force capacity of synthetic molecular motors (2022, April 13)
retrieved 17 April 2022
from https://phys.org/news/2022-04-one-molecule-expose-ability-synthetic-molecular.html
This doc is subject matter to copyright. Aside from any good working for the purpose of private study or analysis, no
aspect may perhaps be reproduced with out the written authorization. The content material is provided for information needs only.